|
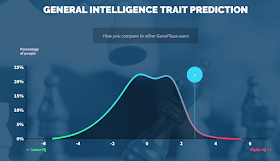
LiveWello report explains how I have a decreased likelihood
of gluten sensitivity and celiac disease |
LiveWello is a site where you can upload your raw DNA data from several testing companies including 23andMe, AncestryDNA, and FamilyTreeDNA and get reports on a number of health related issues. It costs $19.95 to upload your raw DNA data, but they also offer an additional subscription for $5.95 monthly or $60 yearly. But what all do you get with those different options?
With the $19.95 one time upload fee, you gain access to some reports from LiveWello which explain, in plain English, whether you have an increased, normal, or decreased risk of whatever the report is about. In addition, you have access to the "Gene Library" which is a huge library of different health reports created by third parties. It's very extensive, but the drawbacks are that it doesn't explain in plain English what your risks are, and it doesn't allow you to search for a certain type of report. All you can do is scroll through the list of available reports (listed in order that they are added, with no other way to sort them) and hope to find what you're looking for. The reports only provide the raw DNA and leaves the interpretation up to you, and that means you need some understanding of DNA and how genetic variance works. Many of the Gene Library reports don't even tell you what conditions are associated with the gene(s) it's reporting on, suggesting it's only really beneficial for academics or health care professionals.
Disappointing? A bit. For $19.95, you don't get much, and then they want you to pay even more. Compared to options like Promethease, which provides hundreds of reports for only $5 (one time fee), it seems like you're paying a lot more to get a lot less with LiveWello. In addition, note that although you can upload multiple kits to one account, the fees for LiveWello (both the one time upload fee and subscription) apply to each kit you want to upload. So, let's say you manage your kit, and both your parents kits - full access to LiveWello for all of you would cost about $60 in upload fees, and about $18 monthly. That starts to really add up.
 |
Gene Variance report from "Gene Library" template doesn't
include any explanation of what the data means (increased,
decreased, or average risk) |
With the subscription, you gain access to all reports provided by LiveWello, and they also claim you gain access to "all the features included in your gene variance report" (these are for the Gene Library), but I am unclear what this includes since my gene variance reports look exactly the same before and during the subscription. So although the Gene Library is extensive, for most people, it won't be of much use unless you are knowledgeable about gene variance and how they work.
It may look like the color coding explains a tendency towards something good or bad, but it doesn't appear to be consistent and doesn't explain how having both green and red results makes you more or less prone to something.
With the one time fee, you get the following 28 reports (not including Gene Library):
1. COMT Gene and Sensitivity to Pain
2. Nexium, acid reflux disease and CYP2C19 drugs
3. Warfarin and CYP2C9 drugs
4. Plavix, blood thinners and CYP2C19 drugs
5. Response to diabetes medication - Sulfonylureas
6. Hepatitis C Treatment
7. Preference for sweet foods
8. Tramadol and CYP2D6 drugs
9. VDR Taq Gene and Osteoporosis
10. Lupus
11. Vitamin B12
12. MTHFR and Risk of Depression
13. COMT Gene and Personality Traits
14. Vitamin B6
15. Alcohol tolerance
16. Anti-depressant Response: Paxil, Celexa, Effexor or Elavil
17. Folate and the MTHFR gene
18. Caffeine and Anxiety
19. Kidney disease risk and MTHFS gene
20. Oxycodone and CYP2D6 drugs
21. Vitamin A
22. Hormone Replacement Therapy
23. Response to cholesterol lowering medication
24. Sexual Dysfunction due to Celexa, Lexapro, Prozac, Paxil or Zoloft
25. Alcohol abuse and risk of esophageal cancer
26. Zofran and CYP2D6 drugs
27. MAOA - The Warrior gene
28. Metformin
With the additional subscription, you get 93 more reports (not including Gene Library):
1. Aspirin Allergy - Asthma Risk (Subscription only)
2. Risk of Duodenal Ulcer (Subscription only)
3. CYP Gene and Irregular Heart Rhythm (Subscription only)
4. Genes Related to Manic Symptoms in Bipolar Disorder (Subscription only)
5. COMT Gene and Irritable Bowel Syndrome (Subscription only)
6. Histamine Gene and Sensitivity to NSAIDS (Aspirin, Alleve, Advil, Motrin) (Subscription only)
7. Floxacillin Associated Liver Toxicity (Subscription only)
8. Narcolepsy (Subscription only)
9. Susceptibility to Both Crohn's Disease and Ulcerative Colitis (Subscription only)
10. FUT2 Gene and the Gut Microbiome (Subscription only)
11. Hot flashes in post menopausal women (Subscription only)
12. Response to bupropion treatment for smoking cessation (Subscription only)
13. Susceptibility to food poisoning caused by Norovirus (Subscription only)
14. COMT Gene and Antipsychotics (Subscription only)
15. G6PD Gene and Risk of Hemolysis with Bactrim (Subscription only)
16. Methamphetamine and Risk of Psychosis (Subscription only)
17. Painful Menstrual Period and BDNF Gene (Subscription only)
18. COMT Gene and Response to Effexor (Subscription only)
19. MTHFR Gene and Pravastatin Efficacy (Subscription only)
20. Risk of Deep Vein Thrombosis (Subscription only)
21. Levofloxacin and Risk Of Seizures (Subscription only)
22. Response to Treatment of Blood Pressure with Benazepril (Subscription only)
23. Response to steroid treatment of Crohn's disease (Subscription only)
24. COMT Gene and Tobacco Use Disorder (Subscription only)
25. MTHFR Gene and risk of Stroke (Subscription only)
26. Medication Overuse Headaches (Subscription only)
27. Risk of Acute Psychosis with Cannabis (Subscription only)
28. Processed Meat and Risk of Colorectal Cancer (Subscription only)
29. Response to Vitamin E Supplementation (Subscription only)
30. Fat and Obesity (FTO) Gene and Risk of Alzheimer's Disease (Subscription only)
31. Cold Sores (Subscription only)
32. APOE gene and Alzheimer disease risk (Subscription only)
33. Risk of liver damage with Depakote (valproic acid) (Subscription only)
34. Aspirin Allergy - Hives (Subscription only)
35. Risk of Developing Multiple Sclerosis (Subscription only)
36. Efficacy of Blood Pressure Medication, Norvasc (Subscription only)
37. NOS Gene and Response to Viagra (Subscription only)
38. APOE Gene, Cholesterol level and Diets (Subscription only)
39. Vitamin D (Subscription only)
40. Response to Carisoprodol (SOMA) (Subscription only)
41. Heart Failure Treatment with Bidil (Subscription only)
42. Breast Cancer Treatment (Subscription only)
43. MTHFR, Homocysteine and Nitrous Oxide Anesthesia (Subscription only)
44. Hormone Replacement Therapy, SULT1A Gene and Risk of Endometrial Cancer (Subscription only)
45. Abacavir (Subscription only)
46. Migraine response to vitamin supplementation (Subscription only)
47. NTRK2 Gene and Lithium Efficacy (Subscription only)
48. Smoking cessation with nicotine replacement therapy (Subscription only)
49. GNB3 Gene and Antidepressant Toxicity (Subscription only)
50. Response to Narcolepsy Drug - Modafinil (Subscription only)
51. BDNF Gene and Depression - Response to Paxil (Subscription only)
52. Tegretol Efficacy in Treatment of Seizures (Subscription only)
53. NOS3 Gene and Response to Blood Pressure Medications (Subscription only)
54. Phenytoin and CYP2C9 drugs (Subscription only)
55. Heroin Addiction (Subscription only)
56. SLC2A3 Gene and Dyslexia in Children (Subscription only)
57. Sensitivity to Muscle Relaxants Used in General Anesthesia (Subscription only)
58. APOE Gene and Exercise Response (Subscription only)
59. General Anesthesia - postoperative nausea and vomiting (Subscription only)
60. G6PD deficiency, risk of malaria and drug-induced hemolysis (Subscription only)
61. STXBP5L Gene and Facial Aging (Subscription only)
62. Multiple Chemical Sensitivity (Subscription only)
63. MTHFR Gene and Smoking Behavior (Subscription only)
64. Response to Ibuprofen (PTGS gene) (Subscription only)
65. Fibromyalgia and Chronic Widespread Pain (Subscription only)
66. MTHFR Gene and Migraine with Aura (Subscription only)
67. MTHFR, Metformin and risk of blood clots (Subscription only)
68. Tylenol and Risk Of Liver Failure (Subscription only)
69. 5-fluorouracil (Subscription only)
70. MTHFR Gene and Risk of Male Infertility (Subscription only)
71. OPRM1 Gene and Opioid Antagonists (Subscription only)
72. COMT, LRP2 Genes and Risk for Gout (Subscription only)
73. Obesity risk (Subscription only)
74. NSAIDS and Acute Coronary Syndrome risk (Subscription only)
75. SSRI Antidepressants and CYP2C19 Gene (Subscription only)
76. Susceptibility to Hypertension (Subscription only)
77. Hypersensitivity to Mercury (Subscription only)
78. Gluten intolerance genes (Subscription only)
79. Choline - dietary requirement in premenopausal women (Subscription only)
80. COMT Gene and Skeletal Muscle Decline in Older Women (Subscription only)
81. Elite physical power and sprint performance (Subscription only)
82. Omega-3 Fatty Acids (Subscription only)
83. Susceptibility to Pregnancy-induced hypertension (Subscription only)
84. Polycystic Ovary Syndrome (PCOS) (Subscription only)
85. Naltrexone and Treatment of Alcoholism (Subscription only)
86. Earwax and Body Odor (Subscription only)
87. MTHFR gene and Diabetic Neuropathy (Subscription only)
88. Risk of Cough with Blood Pressure Medication (Subscription only)
89. Cluster Headaches and Response to Treatment (Subscription only)
90. COMT Gene and Response to Opiods (Subscription only)
91. Salt sensitivity (Subscription only)
92. Risk of Severe Hypersensitivity to Tegretol (Subscription only)
93. Genes Associated with Empathy (Subscription only)
 |
Not all reports are available depending on which company
you tested with |
Another thing to factor in is the fact that if you uploaded your raw DNA data from AncestryDNA, FamilyTreeDNA, and possibly other companies, some of the reports may not be possible due to the necessary SNPs not being included. Not all testing companies include the same SNPs or the same amount of SNPs. FamilyTreeDNA in particular removes about 3,000 medically relevant SNPs. So you could wind up not even having access to all these reports even when you subscribe. With AncestryDNA, I noticed a number of reports were "incomplete because none of the SNPs in it were found in your raw data." And even if you have some of the SNPs necessary, if you don't have all of them, it still may not be able to generate the report and say you have "insufficient genotypes to determine response for" that report. These included but may not be limited to (and may vary depending on the company you tested with):
1. SLC2A3 Gene and Dyslexia in Children
2. Floxacillin Associated Liver Toxicity
3. Response to diabetes medication - Sulfonylureas
4. G6PD Gene and Risk of Hemolysis with Bactrim
5. Methamphetamine and Risk of Psychosis
6. Tramadol and CYP2D6 drugs
7. Response to steroid treatment of Crohn's disease
8. Oxycodone and CYP2D6 drugs
9. Abacavir
10. NTRK2 Gene and Lithium Efficacy
11. Tegretol Efficacy in Treatment of Seizures
12. NOS3 Gene and Response to Blood Pressure Medications
13. Phenytoin and CYP2C9 drugs
14. Hormone Replacement Therapy
15. G6PD deficiency, risk of malaria and drug-induced hemolysis
16. Response to Ibuprofen (PTGS gene)
17. Sexual Dysfunction due to Celexa, Lexapro, Prozac, Paxil or Zoloft
18. Zofran and CYP2D6 drugs
19. Risk of Cough with Blood Pressure Medication
20. Choline - dietary requirement in premenopausal women
21. General Anesthesia - postoperative nausea and vomiting
22. Vitamin B6
23. Risk of Deep Vein Thrombosis
24. Hot flashes in post menopausal women
25. APOE Gene, Cholesterol level and Diets
26. Nexium, acid reflux disease and CYP2C19 drugs
27. Warfarin and CYP2C9 drugs
28. Plavix, blood thinners and CYP2C19 drugs
29. Narcolepsy
30. Susceptibility to Both Crohn's Disease and Ulcerative Colitis
31. FUT2 Gene and the Gut Microbiome
32. Susceptibility to food poisoning caused by Norovirus
33. Risk of Acute Psychosis with Cannabis
34. Alcohol tolerance
35. Cold Sores
36. APOE gene and Alzheimer disease risk
37. NOS Gene and Response to Viagra
38. Vitamin D
39. Response to Carisoprodol (SOMA)
40. Hormone Replacement Therapy, SULT1A Gene and Risk of Endometrial Cancer
41. APOE Gene and Exercise Response
42. SSRI Antidepressants and CYP2C19 Gene
43. Risk of Severe Hypersensitivity to Tegretol
That's more than a third of all the reports LiveWello provides (not including Gene Library).
Conclusion: The 28 reports you get for $19.95 are probably not worth the money, and can probably be obtained from other venues for less. However, if you're going to spend the $19.95 to upload, you might as well subscribe because it's only another $5.95 and you get a good deal more for that - you can cancel after the first month and you won't be missing anything. If they wind up adding a report you need or want later, you can always renew your subscription for another month. You may want to try cheaper venues like
Promethease first though, you might find it is more comprehensive for much less money. For decent free options, I would try
Codegen,
GeneKnot, or
Impute.
See a list of more places you can upload your DNA to here.